PMOD implements an improved fully parallel 3D thinning algorithm [1]. It uses a 3D binary image obtained by a segmentation method and generates unit-width curve skeletons. The skeletonization algorithm extracts the "center-lines" of an object and uses them to efficiently represent the object. [2] The skeletonization algorithm is complementary to the segmentation.
The curve skeletons are well suited to describe tube-like anatomical structures, e.g. vessels, nerves, and elongated muscles [3].
The steps required to generate e.g. a skeleton for the brain vessels based on an MRI image are illustrated in the capture below:
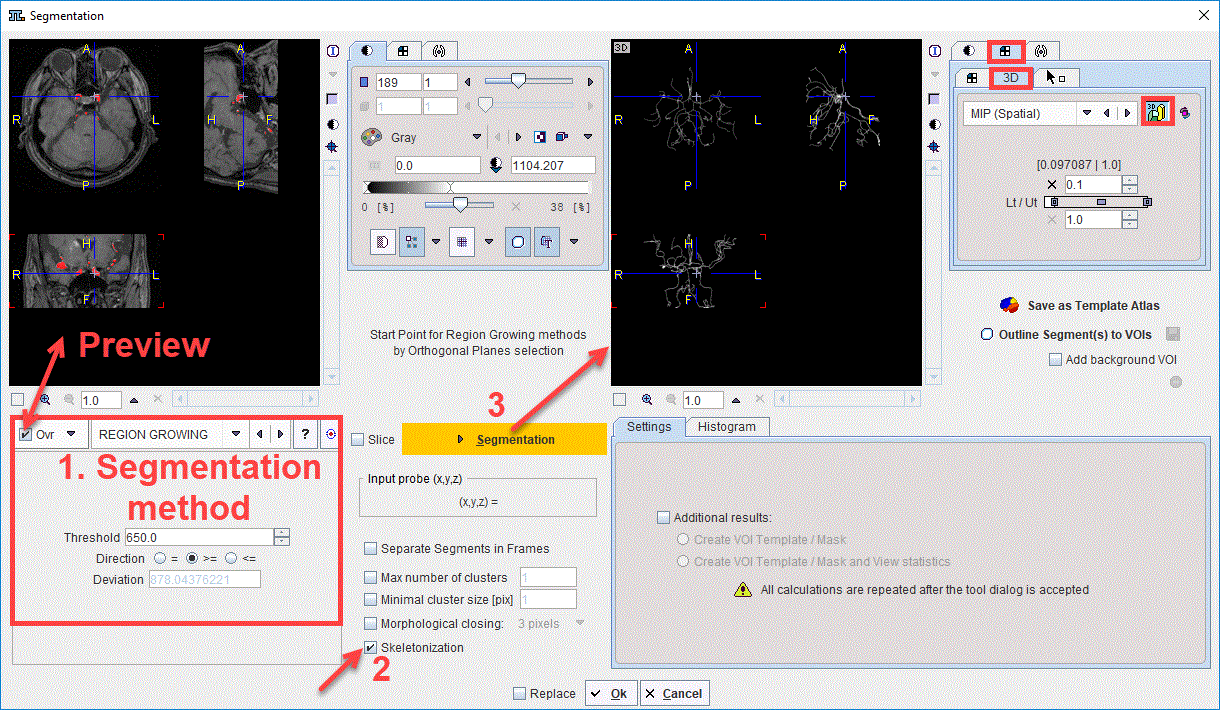
First, the segmentation method is selected in the Segmentation panel. It is recommended to enabled the Ovr button in order to preview the results of the segmentation algorithm in the upper left viewport. Second, the Skeletonization check box is enabled. Finally, the Segmentation button is activated to perform the actual skeletonization of the previewed 3D segment. The result is shown in the upper right viewport. It is recommended to visualize the skeletonization results using the 3D volume rendering layout.
Reference
1.Kálmán Palágyi, A 3D fully parallel surface-thinning algorithm, Theoretical Computer Science 406, October 2008, p. 119–135, DOI
2.Tao Wang, Skeletonization and Segmentation Algorithms for Object Representation and Analysis, PhD Thesis, Spring 2010, Edmonton, Alberta, University of Alberta
3.Tagliasacchi A., Delame T, Spagnuolo M., Amenta N., Telea A, 3D Skeletons: A State-of-the-Art Report, EUROGRAPHICS 2016, Volume 35 (2016), Number 2