The Rigid Matching button provides access to the different types of rigid matching methods. An interface dialog window appears with two tabs. The first one serves to quickly retrieve species sensitive pre-defined or saved configurations of the iterative matching settings, the second one gives access to all parameters.
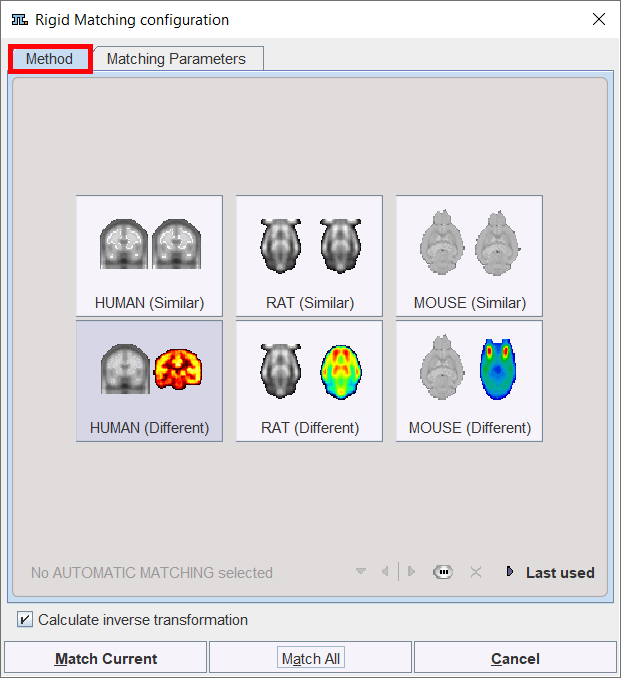
There are predefined methods for intra-modality matching (MRI-MRI, PET-PET), and cross-modality matching. Activating the species and modality corresponding buttons displays the Matching Parameters pane with appropriately configured settings. There are Basic and Advanced parameters on separate tabs.
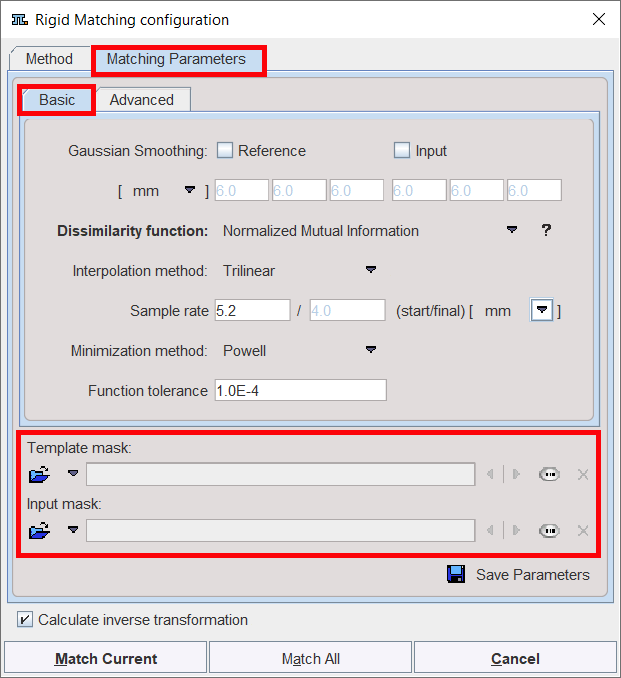
The settings available in the panels allow fine-tuning the basic procedure in a multitude of ways. While there are successful settings (as the predefined ones), experimenting with these configurations may result in improved or faster matches in specific situations.
Basic Parameters
Gaussian Smoothing |
A Gaussian filter with configurable width in mm or pixels can be separately enabled for the Reference and the Reslice study. While this introduces an additional performance burden during start-up, iterations are less likely to get trapped in a local optimum with smoothed images. |
Dissimilarity function |
This is the main definition of the matching algorithm. Note that a short explanation of the selected dissimilarity function can be shown with the ? button besides the selection. The selections are ▪Absolute Difference Sum, and ▪Squared Difference Sum: These are measures based on image subtraction and therefore require images of the same modality. ▪Woods: Partitioned Intensity Uniformity for the registration of MRI-PET images [7], [8]. ▪Mutual Information, Intra- and Cross-Modality: Mutual information (MI) is a term from information theory [1]-[6]. Mutual information can be expressed as the sum of individual entropy terms of the random variables less their joint entropy. MI normalizes the joint entropy with respect to the partial entropies of the contributing signals. The dissimilarity function value is calculated from joint histogram of resampled reference and input data. ▪Mutual Information (PV), Intra- and Cross-Modality: In this MI variant, a partial volume interpolation algorithm is used as a part of the joint histogram construction. The histogram calculations are performed directly on the reference and input data. As a consequence the interpolation method selection in the matching parameters configuration has no relevance for this dissimilarity function. ▪Normalized Mutual Information, Intra- and Cross-Modality: The normalized MI variant also uses partial volume interpolation and additionally a normalization scheme proposed by Studholme [5]. This variant has become very popular in the recent years and performs well in many multi-modality situations. |
Interpolation method |
Type of interpolation used during reslicing. Has an impact on speed, and may also influence convergence. |
Sample rate |
Density of resampling the original images during the matching process. Coarse sampling increases speed dramatically, but too coarse images may not allow any more for accurate matching. 6 or 8 mm is often satisfactory for MRI/PET matching. A strategy with multiple searches can be implemented in combination with the Algorithm runs option: the first matching runs are performed at a coarse resolution, but the last one with a fine sampling rate for an accurate final match. |
Minimization Method |
Powell usually finds the optimal match faster than Downhill Simplex. |
Function tolerance |
Termination criterion for the iterations. |
Template Mask |
Allows defining a mask for the reference image if none was created and/or set to the matching protocol after the reference image was loaded. To discard the mask activate the Clear file or directory button |
Input Mask |
Allows defining a mask for the input image if none was created and/or set to the matching protocol after the input image was loaded. To discard the mask activate the Clear file or directory button |
Save |
Save the parameter settings for later use. |
Calculate Inverse Transformation |
If the box is checked, the inverse transformation is also calculated once the matching completed. |
Advanced Parameters
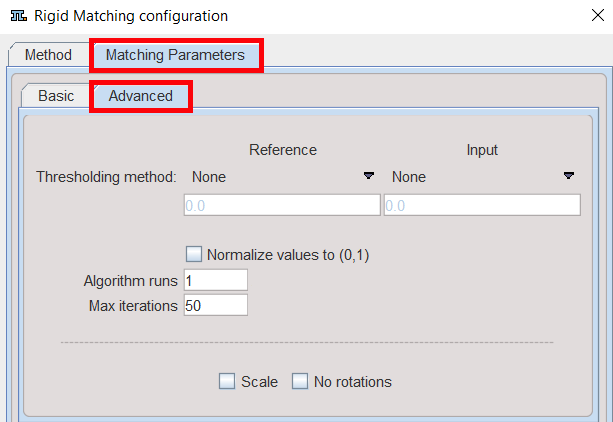
Thresholding method |
The image volume considered during matching can be restricted to a sub-volume by thresholding, eg. by excluding the image background. Absolute values can be defined when User defined option is selected as thresholding method. |
Normalize values to (0,1) |
When this box is checked, the image values are normalized to the numeric range [0,1]. Note that the operation is a scaling, not a binarization of the image. This transformation may be required when applying one of difference criteria, if the dynamic range of the matched images is different for instance because of different administered tracer doses. |
Algorithm runs |
A value > 1 configures multiple successive matching runs, whereby a run is started with the result parameters of the preceding run. |
Max iterations |
A maximal number of optimization steps can be configured to avoid "endless" looping. |
Scale |
If box is checked allows scaling the image during rigid matching |
No rotation |
If box is checked no rotation is performed during the automatic rigid matching. |
Starting Rigid Matching
The Match Current button starts the matching process of the current input series with the given settings. A bar will appear in the status line which is incremented for each function evaluation. With Match all, all Reslice series are sequentially matched.
•The principle of iterative rigid matching
1.An initial set of translation and rotation parameters is assumed or derived.
2.The Reslice study is transformed with these parameters.
3.The match between the Reference and the transformed Reslice study is evaluated according to the given matching criterion (Dissimilarity function).
4.Based on the criterion before and after transformation (better?, worse?), the parameters are modified according to an improvement strategy. The updated parameters are expected to yield a better match.
The steps 2 – 4 are repeated until the improvement drops below a given threshold, the function tolerance.