Preparations
For a simultaneous fit of TACs from multiple studies the user first has to
▪load all of the data sets so that they are available on different tabs;
▪ensure that all of the TACs to be coupled have the same compartment model configuration and the same residual weighting;
▪ensure that all model parameters have reasonable initial values by either fitting the models, or propagating a model configuration.
The coupling is performed on a separate dialog window which can be opened from the Kinetic menu with the Coupled Studies Dialog entry.
Study and Model Selection
The dialog window opens on the first tab Study/Model selection containing three sections.
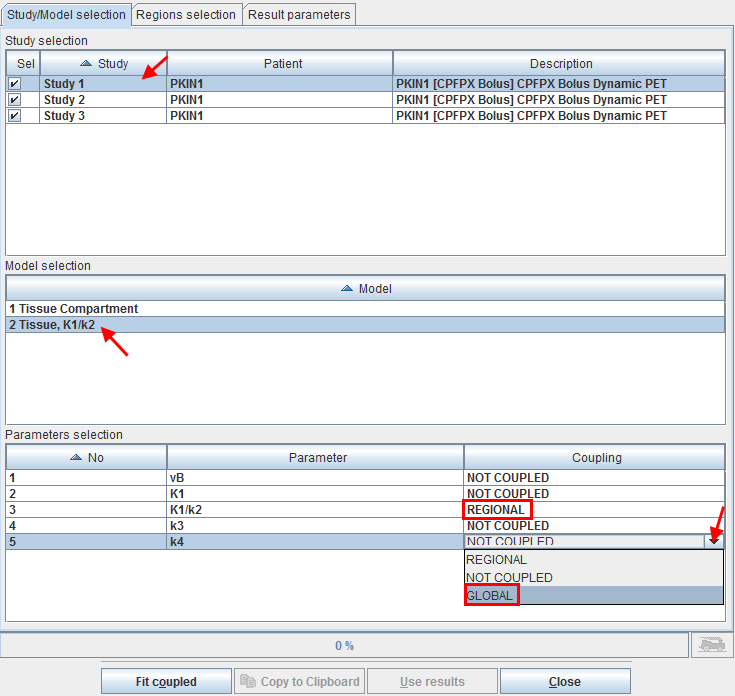
The Study selection lists all open data sets. As soon as one of them is selected in the list, the Model selection list is updated, showing the different models which are used for the TACs of the study. In the example shown above one of the regions (which is not used four coupling) uses a 1 Tissue Compartment model, whereas the regions intended for coupling use the 2 Tissue K1/k2 model.
After selecting the appropriate model, the Parameters selection list is updated showing all parameters of the model together with a Coupling selection. A parameter can have one out of three configurations:
▪NOT COUPLED: The parameter will be individually fitted in the different regional models or kept fix depending on the state of the individual fit box.
▪REGIONAL: The parameter will be coupled among all regions with the same name. For instance, the parameter will have a common value for all caudate regions, a different common one for all putamen regions, etc. Note that with this configuration the state of the individual parameter fit boxes is not relevant, the parameter will be modified in all coupled regions.
▪GLOBAL: The parameter will be coupled among all coupled regions independent of their name. For instance, the parameter will have the same common value among all caudate, putamen, etc regions. Note that with this configuration the states of the individual parameter fit boxes is not relevant, the parameter will be modified in all coupled regions.
Regions Selection and Fitting
The next step is defining the TACs included in the coupled fit on the Regions selection pane as illustrated below.
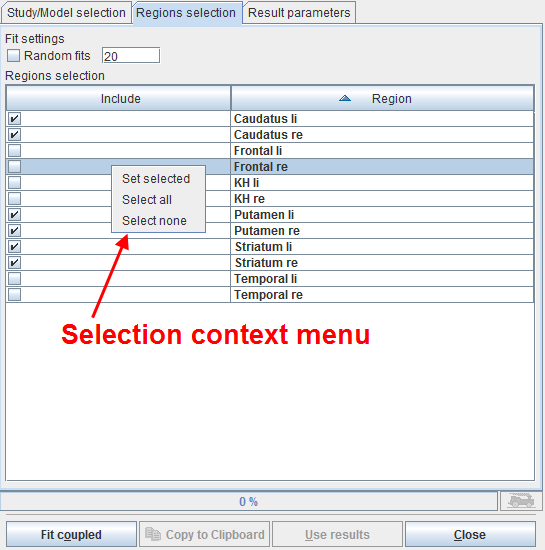
The list contains all regions which are defined in the coupled studies with exactly the same names. Initially all regions are selected for inclusion. Regions can selectively be unselected by checking their box. When working with large number of regions (e.g. from atlas definitions) the context menu may be helpful which can be opened by right clicking into the list. It contains Select all and Select none for quickly selecting or de-selecting all regions, respectively.
Coupled fitting is started using the Fit coupled button and proceeds as follows:
▪A global target curve is created by stacking the TACs of all coupled regions from the selected studies (valid points only).
▪The corresponding weights are calculated according to the weighting configuration for each TAC.
▪A table of fitted parameters is formed by entering the GLOBAL parameters once, the REGIONAL parameters once per region, and adding all the other fit enabled parameters in the coupled regions. The initial values of common parameters are taken from the first study, for the other parameters from the individual regional models. Note that Fit blood parameters has no impact on fitting, all the parameters of the blood curves remain fixed.
The optimizer calculates all model curves, creates the global result curve corresponding to the target curve, weighs all the residuals and forms the global sum of squares as the figure of merit. The parameters are adjusted until the cost function has been minimized. If the box Random Fits is checked, the fitting will be repeated as many times as specified with randomly changed starting parameters and the result with minimal global chi square returned.
Result Parameters
After the coupled fit has completed the resulting parameters in the coupled regions are listed on the Result parameters pane which is immediately shown.

Note in the example above that the REGIONAL parameter K1/k2 has the same value in all regions with the same name, whereas the GLOBAL parameter k4 has the same value in all regions.
The results can be prepared for use in another program such as MS Excel with the Copy to Clipboard button. The Use results button transfers the result parameters together with their standard errors to the individual models and closes the dialog window, whereas the results are discarded by the Close button.