The PKIN models can be applied as part of a processing pipeline. For instance, a dynamic PET series can be motion corrected, an atlas fitted to the data and the regional TACs calculated which are then fitted by a model.
Association of Blood Components to Dynamic PET
For blood-based models the related information such as input curve, whole blood curve, and the parent fraction of an dynamic PET must be specified. This requires that the data resides in a PMOD database and uses the "association" concept which allows defining relationships between data elements.
For instance, the whole-blood curve is specified as follows. In the Patients list of the database browser select the patient, and in the Series list the dynamic PET. Next, activate the association list as illustrated below and select the Associate Whole blood entry.
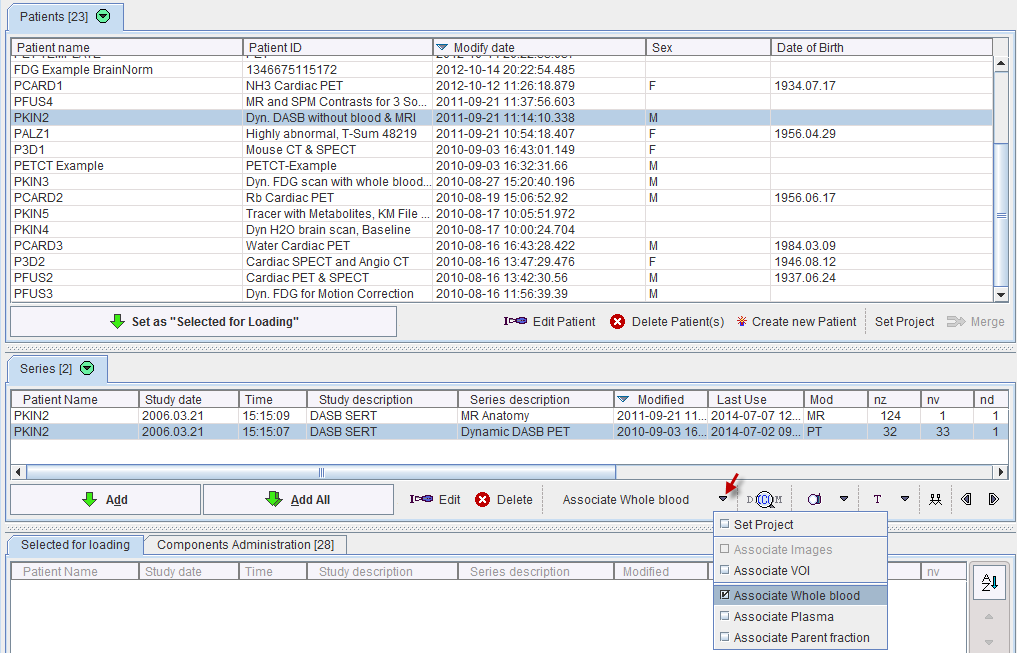
A component browser appears which is used to select the whole-blood curve. The same procedure is repeated for the input curve with the Associate Plasma entry, and if applicable with the Associate Parent fraction. From then on the roles of the blood curves is established, and selection of the dynamic PET is sufficient for modeling purposes.
Kinetic Modeling External Tool
Please refer to the PMOD Base Tool Users Guide for general explanations about setting up batch pipe processing. A modeling stage can be included by means of the Kinetic Modeling external tool which has the interface illustrated below.
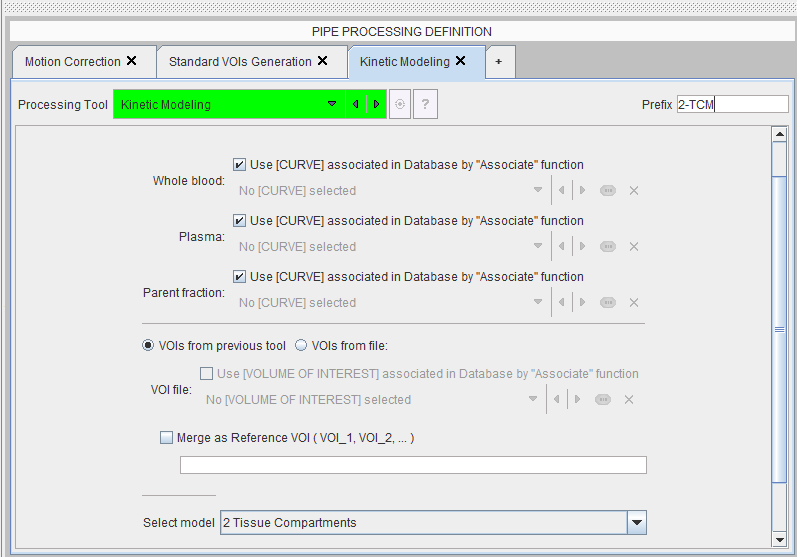
For blood-based models the whole-blood, plasma and parent fractions have to be specified. Unless data of the same study is processed multiple times with the same blood data, the associations have to be used for a general specification. With VOIs from previous tool the VOI definitions produced by the pipeline will be applied. Again, VOIs from file can only be applied wen processing the same data multiple times, or if the images have been normalized to a standard space.
For reference tissue models the reference TAC can be generated by combining a set of VOIs. The set has to be specified in a comma-separated list such a Cerebellum_l,Cerebellum_r, or using wildcards such as Cerebellum*.
Finally, the tissue model to be fitted to each regional TAC can be chosen from the Select model list.