After successful normalization the mapping between the Input and the atlas space is established. Consequently, the brain structures which are defined in the atlas space can be mapped to the Input subject space and shown in the overlay.
Calculate the Brain Segments
Activate the Segment Brain action button at the right bottom of the NORMALIZED layout to prepare the images in the two spaces. As a result, the (averaged) Input image is overlaid with an image of the transformed atlas regions called BRAIN SEGMENTS.
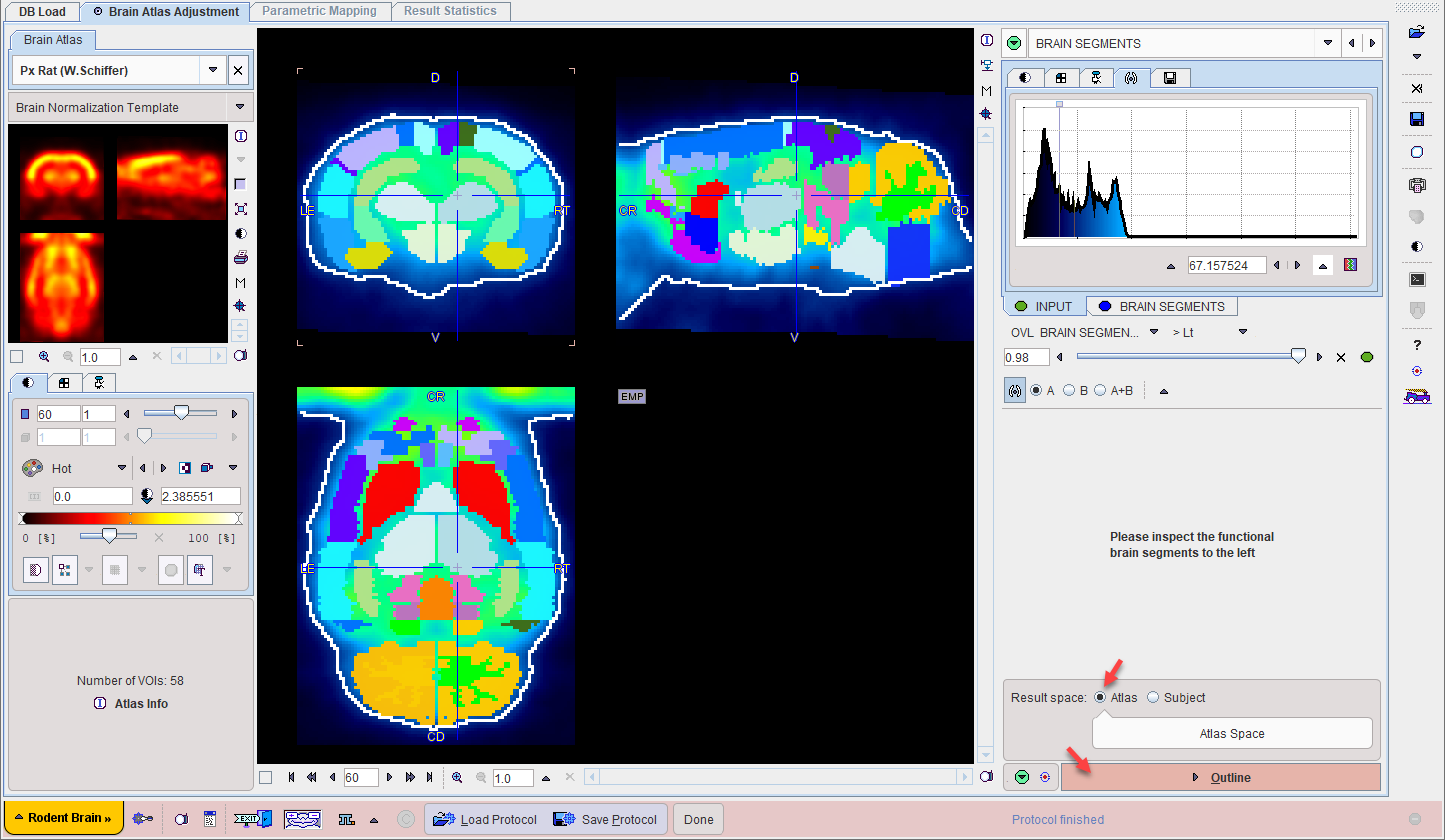
Please perform a final validation of the region placement. As mentioned before, a certain degree of deviation can't be avoided and needs to be handled by adjusting the generated brain VOIs.
Result Space Definition
There are two image spaces where the results can be generated and the statistics calculated:
1.Atlas space: The Input image is transformed to the space of the atlas. This option is preferable for pooling the resulting images (functional image, parametric map) of a group of subjects and performing an analysis such as SPM.
2.Subject: The atlas brain VOIs are transformed to the Input space. This option has the advantage that the statistics are calculated on the original Input data, whereas in the atlas space the data has been resampled.
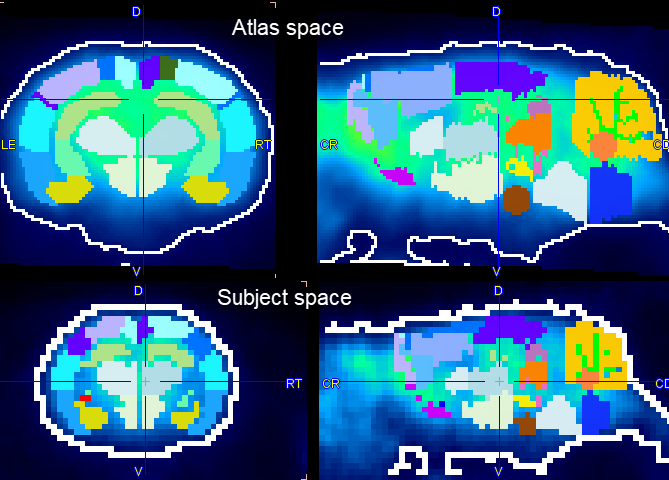
The information visualized on the page is updated as soon as the Result space configuration is changed. The illustration below shows the results when selecting the two spaces. Clearly, the PET Input series has a much lower resolution than the atlas and correspondingly the segments appear heavily pixelated.
Brain Structure Outlining
Once the result space has been specified, the brain structures are fully defined and can be outlined to create contour VOIs. This process is started with the Outline action.