The Learning Set was exported as an R workspace and transferred to cloud computing infrastructure for training. PMOD can be installed on the virtual machine provided by the cloud computing provider and training launched using the R workspace:
The parameters used were:
•4 GPU V100, memory 488 GB
•382 samples (305 training, 77 validation)
•pixel size 0.5 x 0.5 x 0.5 mm
•batch size 16
•Learning rate 0.05
•epochs 300
Training took 3 hours 8 minutes at 0.123 seconds/sample. The loss values for training and validation were extracted from the Manifest and plotted:
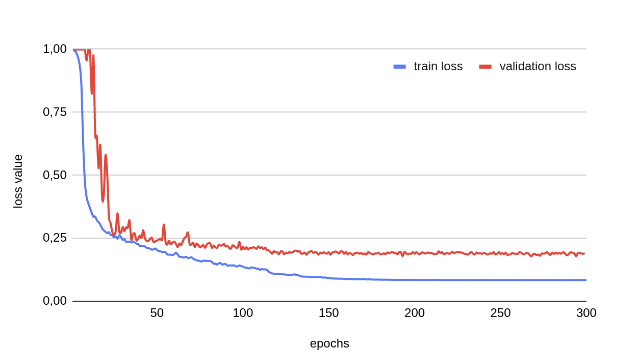
The plot illustrates how the loss value reduces with each epoch until a plateau is reached.
The Learning Set, Weights and Manifest were exported from the virtual machine and used to create a new model folder Rat Brain Dopaminergic PET in the weights subfolder of the Multichannel Segmentation folder in Pmod4.2/resources/pai. After this Deployment it was possible to test the model performance of comparable rat brain [11C]-methylphenidate data for which an anatomical MR image was available.
The resulting VOIs are shown below on the EARLY, LATE and anatomical reference MR images:
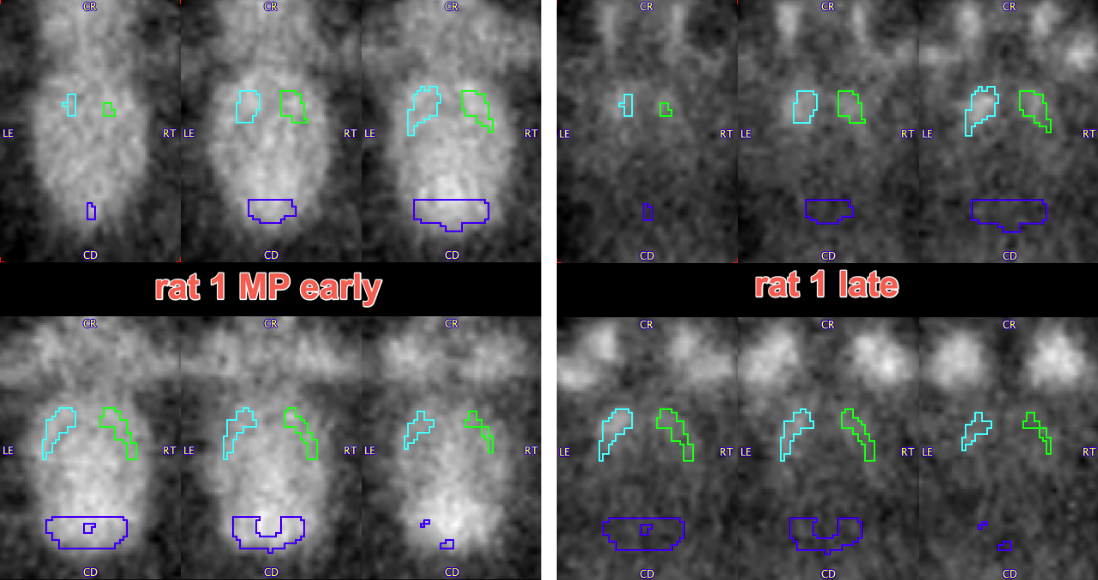
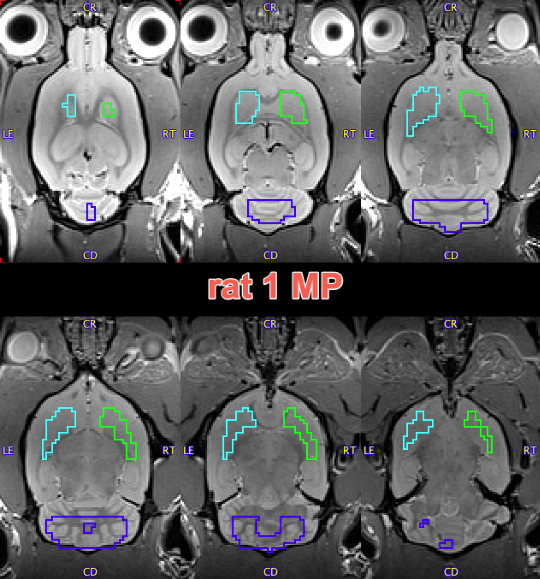
Time-activity curves were extracted from the dynamic PET series using both these VOIs and reference VOIs from PNROD. The two sets of TACs are shown below with matched axes:
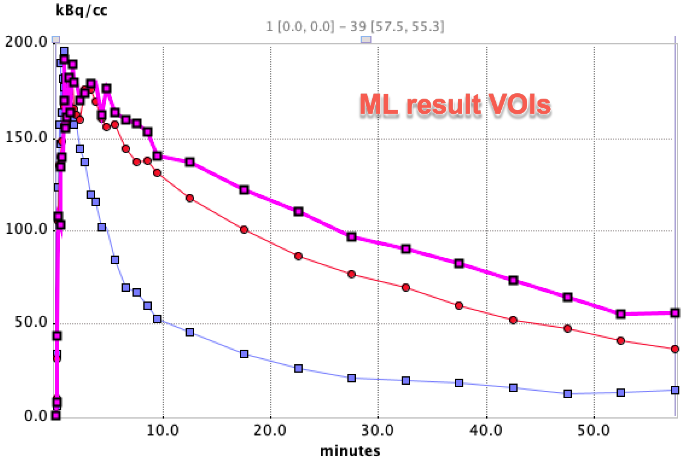
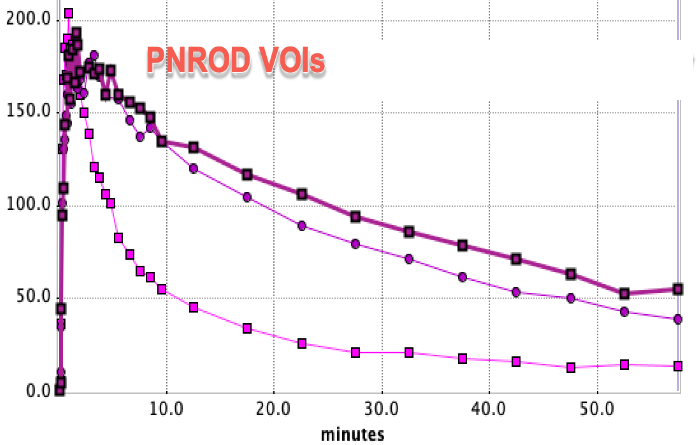
The curve with faster washout in each case is the cerebellum.