The Perfusion (pCASL MRI) model provides an implementation of the data processing for ASL (Arterial Spin Labeled) data described in a recent consensus paper by Alsop et al. [1]. ASL perfusion MRI permits noninvasive quantification of blood flow. With current state-of-art approaches high quality whole-brain perfusion images can be obtained in a few minutes of scanning.
ASL uses arterial blood water as an endogenous diffusible tracer by inverting the magnetization of the blood using RF pulses. After a delay to allow for labeled blood to flow into the brain tissue, "labeled" images are acquired that contain signal from both labeled water and static tissue water. Separate "control" images are also acquired without prior labeling of arterial spins, and the signal difference between control and labeled images provides a measure of labeled blood from arteries delivered to the tissue by perfusion. The lifetime of the tracer is governed by the longitudinal relaxation time of blood, which is in the range of 1300–1750 ms at clinical field strengths [1].
Perfusion is calculated using the following formula (upper equation on p15 of [1]):
![]()
λ is the brain/blood partition coefficient, SIcontrol and SIlabel are the signal intensities in the control and label images respectively, T1, blood is the longitudinal relaxation time of blood in seconds, α is the labeling efficiency, SIPD is the signal intensity of a proton density weighted image, τ is the label duration, PLD the post label delay. The factor of 6000 converts the units from ml/g/s to ml/(100g)/min, which is customary in the physiological literature. Surround subtraction was applied according to Warnock et al. [2].
Acquisition and Data Requirements
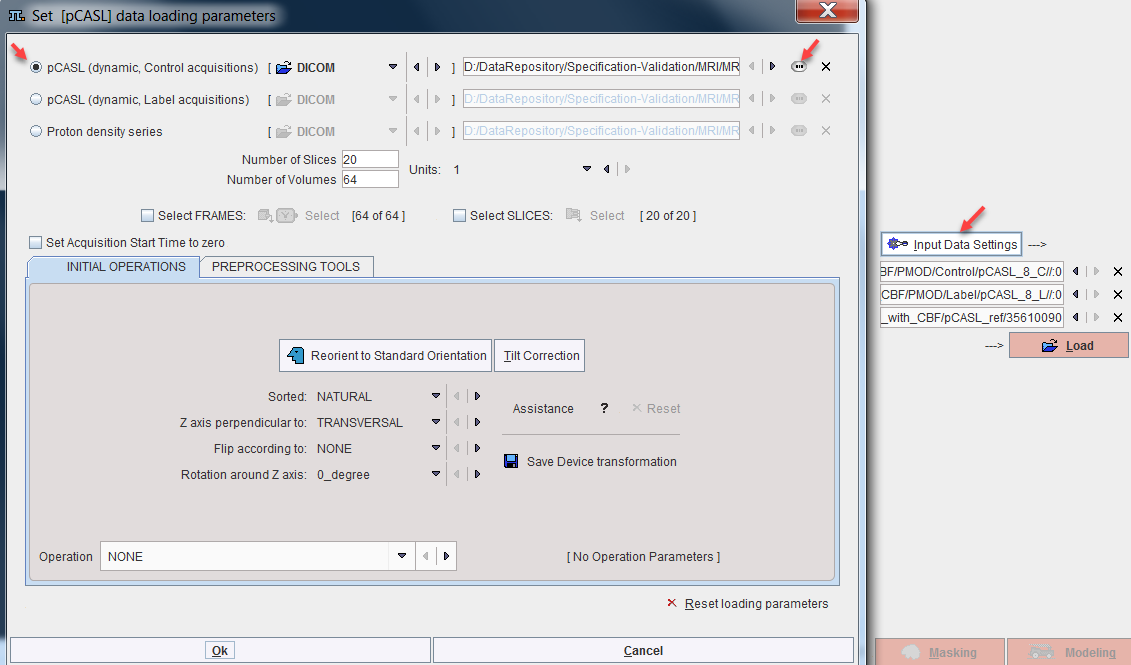
It is recommended [1] to acquire control and label scans in an alternating fashion. However, for import into the Perfusion (pCASL MRI) model the control and label images must be organized as two separate (dynamic) series. Additionally, a matched proton density weighted image is also required. The three data sets can be configured as illustrated below. Select the Input Data Settings to open the data loading dialog window. Configure the "control" series by selecting the pCASL (dynamic, Control acquisitions) radio box, setting the appropriate data format, and selecting the dat with the button indicated to the right. Proceed similarly for the "label" and the proton density series.
Model Preprocessing
When arriving at model preprocessing, manually enter the parameters of the acquisition and the tissue properties. No information is extracted from the image header. However, the information is serialized and will be conserved when using the model repeatedly.
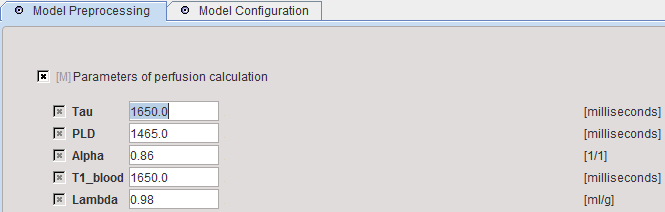
Tau |
Duration of label pulse [msec]. |
PLD |
Post labeling delay [msec]. |
Alpha |
Labeling efficiency. |
T1_blood |
Longitudinal relaxation time of blood at the magnetic field strength used [msec] |
Lambda |
Brain/blood partition coefficient of water [ml/g] |
Model Configuration
Only one parametric map can be generated
![]()
F |
Average perfusion during the acquisition (average of Perfusion(t)). |
However, pixelwise calculation creates an additional results which can be found in the list of parametric maps.
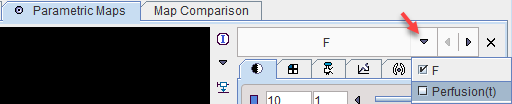
Perfusion(t) |
Dynamic series representing perfusion at the time of the individual acquisitions. |
Reference
1.Alsop DC, Detre JA, Golay X, Gunther M, Hendrikse J, Hernandez-Garcia L, Lu H, MacIntosh BJ, Parkes LM, Smits M et al: Recommended implementation of arterial spin-labeled perfusion MRI for clinical applications: A consensus of the ISMRM perfusion study group and the European consortium for ASL in dementia. Magn Reson Med 2015, 73(1):102-116. DOI
2.Warnock G, Ozbay PS, Kuhn FP, Nanz D, Buck A, Boss A, Rossi C: Reduction of BOLD interference in pseudo-continuous arterial spin labeling: towards quantitative fMRI. J Cereb Blood Flow Metab 2017:271678X17704785. DOI