The 2-Tissue Compartment, K1/k2 model implements fitting a two-tissue compartment model in each image pixel. However, because of the high noise level in pixelwise TACs the fitting of a full 2-tissue compartment model is often not successful. One way to alleviate this problem is to reduce the number of fitted parameters, for instance by fixing the values of k4 and/or K1/k2 at a value which can be reasonably assumed as constant across all tissues. As an example, with PMP, Koeppe et al. [1] fixed k4 at a value of zero and the distribution volume of the non-displacable compartment K1/k2 at a value determined beforehand with a regional TAC analysis.
Processing is done in the following way:
During Preprocessing a TAC is read from a file or averaged in a specified VOI. The TAC is then iteratively fitted to a 2-tissue compartment model which is described by the parameters K1, K1/k2, k3, k4. Each of the parameters can be estimated or, alternatively, fixed at a value which is known a priori. With PMP, for example, k4 was fixed at a value of zero. K1/k2 represents the distribution volume of non-specific binding. It is used as a fit parameter instead of k2 because often K1/k2 can be assumed to be identical in tissues with and without specific binding.
During model-processing, the same 2-tissue compartment model is fitted to the TAC in each individual image pixel. The parameters resulting from the Preprocessing fit are used per default as the starting values of the pixelwise iterative fits. These values, however, can be modified in the model parameters dialog, as well as the fitting flags. The default behavior is suitable for a tracer such as PMP: K1, and k3 are fit-enabled, while K1/k2, is fixed.
Note that iterative fitting in all image pixels is a computationally intensive process. For efficiently working with this model it is recommended:
▪Use a computer system with multiple processing cores.
▪Define a suitable mask.
▪First test the model configuration by processing only the current slice by enabling the corresponding box in the taskbar ![]() .
.
▪Set up a batch and run the fitting over night.
Note: By fixing k3 and k4 at a value of 0 the 2-Tissue Compartment, DV model can be used to fit a 1-tissue compartment model.
Acquisition and Data Requirements
Image Data |
A dynamic PET data set with 2-tissue kinetics. |
Blood Data |
Input curve from the time of injection until the end of the acquisition. Optionally whole-blood activity can be used for spillover correction. |
TAC1 |
A time-activity curve (or VOI) of representative tissue used to determine the starting parameters and the K1/k2 ratio. |
Blood Preprocessing
It is assumed that no Preprocessing other than an optional decay correction must be applied to the plasma activity. This blood data serves as the input curve of the 2-tissue compartment model.
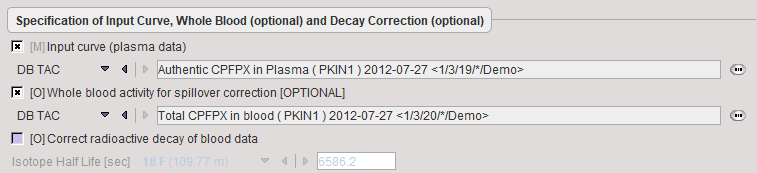
Model Preprocessing
The Preprocessing dialog specifies a tissue time-activity curve (TAC1, FILE or VOI), optionally the total blood activity for spillover correction, and in the Preprocessing parameters a 2-tissue compartment model configuration. The model is fitted to the TAC during Preprocessing and the values updated accordingly.
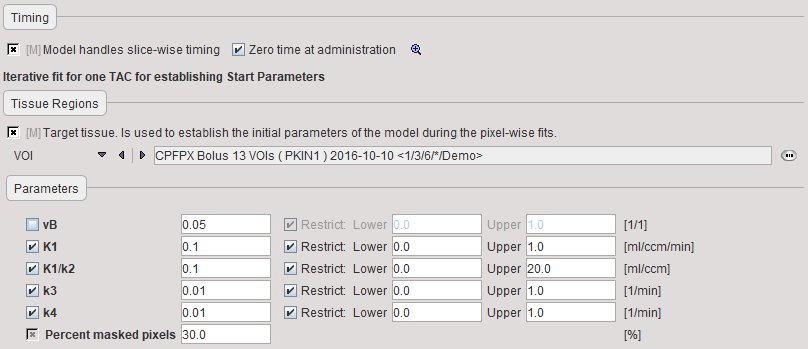
vB |
Blood volume fraction defining the pixelwise blood spillover correction. |
K1,K1/k2,k3, k4 |
Rate constants of the 2-tissue compartment model. |
Percent masked pixels |
Exclude the specified percentage of pixels based on histogram analysis of integrated signal energy. Not applied in the presence of a defined mask. |
Note that the user must initially provide reasonable starting values for the fit-enabled parameters. They are most easily obtained in the PKIN tool. The parameters will be adjusted after model preprocessing and serving as the initial values of pixelwise fitting.
Important: The values and the fit flag of the parameters will be used in pixelwise fitting. If K1/k2 is fitted in pre-processing but should be fixed for the pixelwise fits, return to the Model Preprocessing tab, remove the K1/k2 fit flag, and move forward again with the redt buttons in the lower right.
Model Configuration
The model dialog contains the parameters of the 2-tissue compartment model. Maps can only be generated for parameters which have been enabled for fitting in the Model Preprocessing panel, because the others remain fix for all pixels.
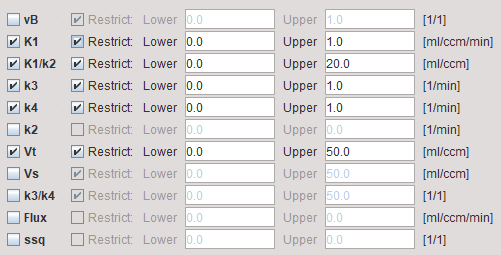
vB |
Blood volume fraction defining the pixelwise blood spillover correction. To fit, please activate in the Preprocessing tab. |
K1,k2,k3, k4 |
Rate constants of the 2-tissue compartment model. |
K1/k2 |
Distribution volume of the non-displaceable compartment. This is a fitting parameter, while k2 is calculated from K1 and K1/k2. |
Vt |
Distribution volume. |
Vs |
Distribution volume of the second compartment. It is only defined for a reversible configuration where k4 has been checked for fitting. |
k3/k4 |
Binding potential of receptor tracers. |
Flux |
Flux to the second compartment: Flux = (K1k3 )/(k2+k3) |
ssq |
Sum-of-squares of the resulting fit. This map can be used to isolate regions with poor fits. |
Reference
1.Koeppe RA, Frey KA, Snyder SE, Meyer P, Kilbourn MR, Kuhl DE: Kinetic modeling of N-[11C]methylpiperidin-4-yl propionate: alternatives for analysis of an irreversible positron emission tomography trace for measurement of acetylcholinesterase activity in human brain. J Cereb Blood Flow Metab 1999, 19(10):1150-1163. DOI