The 1-Tissue (Zhou GRRSC) model implements fitting a 1-tissue compartment model in each image pixel. It is based on a multi-linear formulation of the operational equation, which can be fitted by a fast and reliable weighted linear regression (WLR) method. To improve the signal-to-noise ratio in the calculated parametric maps Zhou et al. [1] have extended the method by ridge regression (RR). In short, the parametric map calculation performs the following steps:
1.A WLR fit is performed for the TAC in each image pixel.
2.The resulting parametric maps of vB, K1 and k2 are then spatially smoothed.
3.A ridge factor is calculated for each pixel using the smoothed parametric maps and the estimated noise variance (difference between signal and fit). It is proportional to the noise.
4.The cost function is extended by a penalty term which is driven by the ridge factor. The noisier a pixel, the higher the penalty.
5.Ridge regression estimates the optimal parameter set vB, K1, k2 a for the penalized cost function. The noisier a pixel, the more will the solution tend towards the smoothed parametric map of the WLR step.
Implementation details of the 1-Tissue (Zhou GRRSC) model:
•The weighted linear regression and the ridge factor calculation are performed during the PXMOD Preprocessing step, whereas the ridge regression runs during the pixelwise processing.
•The Generalized Ridge regression with Spatial Constraint variant of ridge regression described by Zhou et al [1] is implemented which supports spatially varying ridge factors.
•Multi-linear fitting employs the singular value decomposition (SVD) method, using the frame durations as weighting factors.
•The operational equation (16) in [1] has been re-written to accomodate the geometrical variant of the operational equation:
![]()
•The blood volume fraction vB can be fixed at a certain value, or fitted in each individual pixel.
•The smoothness of the result maps is determined by the width of the smoothing filter.
Acquisition and Data Requirements
Image Data |
A dynamic PET data set. |
Blood Data |
Input curve from the time of injection until the end of the acquisition. |
Blood-related TAC |
Optional. The delay and dispersion fit can be applied for brain perfusion data with [15O]-H2O scans. |
Blood Preprocessing
The same blood Preprocessing steps are available as for the brain perfusion model, but except for the data definition field all options are initially disabled as shown below.
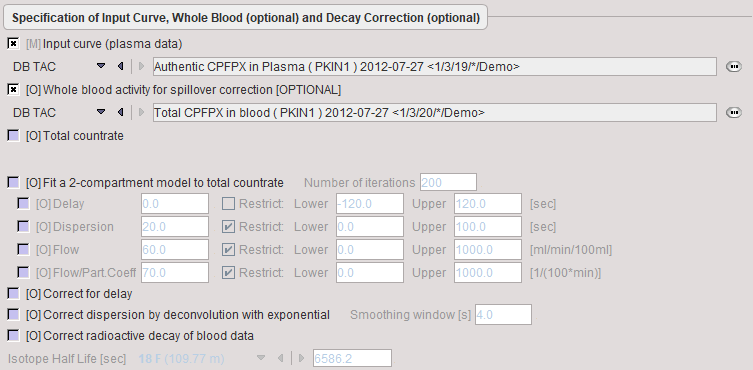
If no whole-blood TAC is defined, the plasma curve will be used for spillover correction.
Model Preprocessing
During model Preprocessing a look-up table is calculated within a range of k2 values. The specifications include an optional TAC which is interpreted as whole-blood activity to be subtracted from the pixelwise TACs.
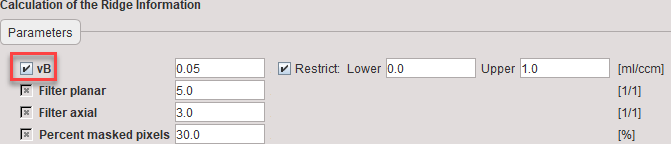
vB |
Blood volume fraction defining the pixelwise blood spillover correction. |
Filter planar |
Number of pixels in the smoothing filter in x and y. |
Filter axial |
Number of pixels in the smoothing filter in z. |
Percent masked pixels |
Exclude the specified percentage of pixels based on histogram analysis of integrated signal energy. Not applied in the presence of a defined mask. |
Model Configuration
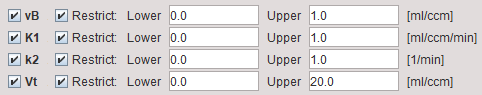
vB |
Blood volume fraction defining the pixelwise blood spillover correction. To fit, please activate in the Preprocessing tab. |
K1 |
K1 rate constant of 1-tissue compartment model. |
k2 |
k2 rate constant of 1-tissue compartment model. |
Vt |
Distribution volume. |
Reference
1.Zhou Y, Huang S, Bergsneider M. Linear Ridge Regression with Spatial Constraint for Generation of Parametric Images in Dynamic Positron Emission Tomography Studies. IEEE Tans Nucl Sci. 2001;48(1):125-130.